Spark Distributed ML model with Pandas UDFs
Why?
Recently, I am learning about Apache Spark distributed computing platform that executes Machine Learning tasks for Big-Data system. Spark has 2 languages used, scala and python (pypark), so this article refers to Pyspark, maybe I will write an article about scala (not today 😍). The full code can be found at the repo Github. Vietnamese version can be read at Vie.
The problem is that I have a super large data file about dogs and cats that need to be filtered into two separate places, to do that, I use the god object detection CNN model mentioned in the previous post Serving ML Models in Production with FastAPI and Celery train again with Dataset. Model training guide here Cats vs Dogs Classification using CNN Keras to classify dog and cat images but instead of checking each image and classifying, process in batch form and use spark to distribute it to many worker nodes to test the processing ability. big data management in spark and make a fun benchmark of python, udf, and pandas udf.
After a while and circling around, charging :v (searching) up and down with google, I found some of the following concepts that may be useful 🔥.
1. UDF & PySpark UDF ?
UDF: User Defined Functions, if you are already using SQL, then UDF is nothing new to you as most traditional databases support user defined functions.
Python UDF’s: is similar to UDF on database. In PySpark you create a function in Python syntax and wrap it with PySpark SQL udf() or register it as udf and use it on DataFrame and SQL respectively. Simply put, write a function in python and use it to act on the dataframe. eg have a dataframe with column X as float and want to add 1 to each element of column X using udf in pyspark.
from pyspark.sql.functions import udf
@udf('double')
def plus_one(x):
return x + 1
#(ss là spark Session)
df = ss.range(0, 3).withColumn('id', (col('id')).cast('integer')).withColumn('v', rand())
df.show()
@udf('double')
def plus_one(v):
return v + 1
df.withColumn('v', plus_one(df.v)).show()
+---+--------------------+ +---+--------------------+
| id| x| | id| x|
+---+--------------------+ +---+--------------------+
| 0| 0.1| -> | 0| 1.1|
| 1| 0.2| | 1| 1.2|
| 2| 0.3| | 2| 1.3|
2. Pandas UDFs ?
Called vectorized UDF — Basically, A Pandas UDF is a UDF that uses Apache Arrow to transfer data and uses Pandas to process that data. To visualize what Apache Arrow is, you can see the picture below (internet source). Apache arrow specifies a standardized language-independent column-memory format for flat and hierarchical data, organized for efficient analytic operations on modern hardware.
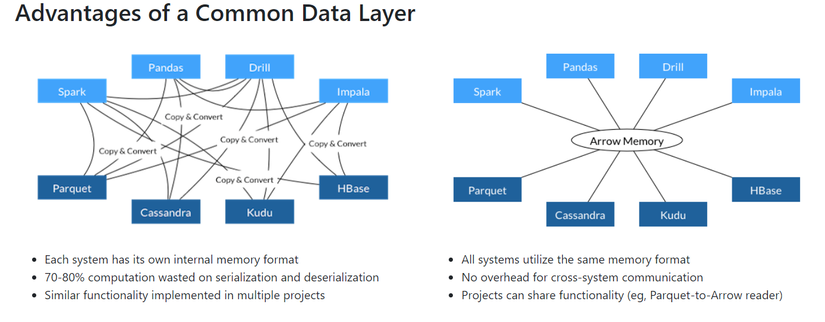
Apache Arrow takes advantage of column caching to reduce IO and speed up analytical processing performance (internet source).

=> Pandas UDF allows vectorized operations that can increase performance up to 100 times compared to Python UDFs row-by-row. In Pyspark, there are 3 types of Pandas UDF: Scalar, Grouped Map and Grouped Aggregate. I will mention Scalar. Is a UDF that converts the Pandas Series into a Pandas Series, where the length of the returned Pandas Series must be equal to the length of the first Pandas Series.
3. Choice ?
So when to use udf and when to use pandas udf:
- Udf does all its operations on a single Node while Pandas udf distributes data to multiple Nodes for processing.
- Unless your data is large enough that it cannot be processed by just one spark node then you should use pandas udf.
- If the data is too small or the task to be performed is not too complex, the performance of udf may be better than Pandas udf.
Distributed ML model with Pandas UDFs
Going back to the original problem of separating data with images of dogs and cats from the dataset. First proceed to clone the repo Github
# clone repo and start jupyter-lab
git clone https://github.com/dnguyenngoc/lab-spark.git \
&& cd lab-spark \
&& docker-compose up
The structure of the project is as follows:
├── spark
├── jupyterlab
├── share_storages
├── lab
├── data
├── images
├── models
├── docker-compose.yaml
...
After executing the docker-compose up command, the services will start. It can take a while before everything is up and running. Note you can customize spark configuration to suit your computer from docker-compose.yaml
| Service | URL | Password |
|---------------|------------------------|----------------|
| Lab | http://localhost:8888 | 1q2w3e4r |
| Spark Master | http://localhost:8080 | None |
Go to http://localhost:8888 and use the password 1q2w3e4r to log in. The notebook containing the entire code for this article is located at Distributed ML model with Pandas UDFs.ipynb. Explain the code a bit. The first is Create session connect to spark cluster with calling spark worker nodes with 2 CPU cores and 2GB RAM configuration.
ss = SparkSession.builder.master('spark://spark-master:7077') \
.appName("test") \
.config("spark.executor.memory", "2g") \
.config("spark.driver.memory", "2g") \
.config("spark.executor.cores", "2") \
.config("spark.driver.cores", "2") \
.getOrCreate()
sc = ss.sparkContext
sc
Show what data you have:
paths = glob.glob('/usr/local/share_storages/data/image/dog-cat/*.jpeg')
rows = 2
plt.figure(figsize=(14,7))
for num, x in enumerate(paths):
image = PIL.Image.open(x)
plt.subplot(rows,3, num+1)
plt.title(x.split('/')[-1])
plt.axis('off')
plt.imshow(image)
plt.show()

Create a dataset of 3000 images from a few original images because I only have 3 cats and 3 dogs, so I have to fake to have a large data file… :v
X1 = [100, 500, 1000, 1500, 2000, 2500, 3000]
random_path = [random.choice(paths) for i in range(3000)]
df = pd.DataFrame(random_path, columns=['path'])
df['label'] =df['path'].apply(lambda x: x.split("/")[-1].split(".")[0]
)
pdf = [df.iloc[: x] for x in X1[:-2]]
# pdf = [df.iloc[: x] for x in X1]
sdf = [ss.createDataFrame(df.iloc[: x]) for x in X1]
print(pdf[0].iloc[: 3])
sdf[0].show(3)
Define function preprocessing image:
IMAGE_SIZE = 192 # (image input 192x192)
BATCH_SIZE = 2
# Preprocess an image
def preprocess_image(image):
image = tf.image.decode_jpeg(image, channels=3)
image = tf.image.resize(image, [IMAGE_SIZE, IMAGE_SIZE])
image /= 255.0 # normalize to [0,1] range
return image
# Read the image from path and preprocess
def load_and_preprocess_image(path):
image = tf.io.read_file(path)
return preprocess_image(image)
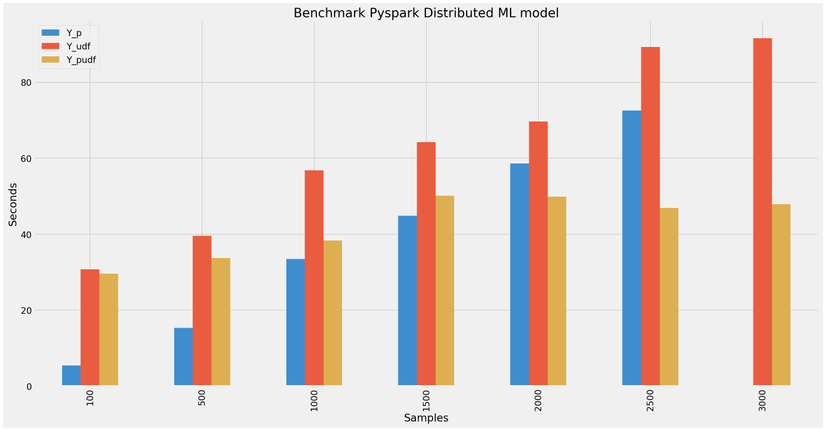
To analyze the processing speed of python, pyspark udf and pandas udf, create a sample file consisting of [100, 500, 1000, 1500, 2000, 2500, 3000] samples from the dataframe, respectively. created. Then Calculate the execution time of python with 4 CPU cores and 4GB RAM and pyspark udf and pandas udf with 2 nodes spark 2 core CPU, 2GB RAM each. Doing the first turn is pure python loading the model and predicting (I set batch_size predict to 2).
X_p = []
X_d = []
def process_batch_python(all_paths):
start_time = time.perf_counter()
data_lenght = len(all_paths)
model = keras.models.load_model("/usr/local/share_storages/data/model/dog_cat.h5")
preprocessed_imgage = np.array([load_and_preprocess_image(item) for item in all_paths])
predictions = model.predict(preprocessed_imgage, batch_size=BATCH_SIZE)
predicted_labels = [{"class": "Cat", "score": x[0]} if x[0] >= 0.5 else {"class": "Dog", "score": x[0]} for x in predictions]
end_time = time.perf_counter()
return predicted_labels, end_time - start_time
for x in X1:
pred, count_time = process_batch_python(df.iloc[:x]['path'])
X_p.append(count_time)
X_d.append(pred)
print("{} sample: {}s".format(x, count_time))
100 sample: 5.428100499997527s
500 sample: 15.379066599998623s
1000 sample: 33.48210130000007s
1500 sample: 44.866816599998856s
2000 sample: 58.65093120000165s
2500 sample: 72.51571390000026s
3000 sample: ngỏm.
Similar to Pyspark udfs:
X_udf_d = []
X_udf_d = []
def process_batch_udf(sdf):
start_time = time.perf_counter()
model = keras.models.load_model("/usr/local/share_storages/data/model/dog_cat.h5")
bc_model_weights = sc.broadcast(model.get_weights())
def cover_model():
model.set_weights(bc_model_weights.value)
return model
model_fn = cover_model()
@udf(StringType())
def predict(img):
processed_images = np.array([load_and_preprocess_image(img)])
predictions = model_fn.predict(processed_images, batch_size=BATCH_SIZE)
predicted_labels = ["Cat,{}".format(x[0]) if x[0] >= 0.5 else "Dog, {}".format(x[0]) for x in predictions]
return predicted_labels[0]
x= sdf.withColumn('v2', predict(sdf.path))
x.show(1)
end_time = time.perf_counter()
return sdf, end_time - start_time
for x in range(len(X1)):
pred, count_time = process_batch_udf(sdf[x])
X_udf.append(count_time)
X_udf_d.append(pred)
print("{} sample: {}s".format(X1[x], count_time))
100 sample: 30.738601299999573s
500 sample: 39.60790839999754s
1000 sample: 56.82039040000018s
1500 sample: 64.21761259999766s
2000 sample: 69.66648029999851s
2500 sample: 89.24947419999808s
3000 sample: 91.57887720000144s
Finally the Pandas Udf:
X_pudf = []
X_pudf_d = []
def process_batch_pandas_udf(sdf):
start_time = time.perf_counter()
model = keras.models.load_model("/usr/local/share_storages/data/model/dog_cat.h5")
bc_model_weights = sc.broadcast(model.get_weights())
def cover_model():
model.set_weights(bc_model_weights.value)
return model
def keras_model_udf(model_fn):
"""
Wraps an Keras model into a Pandas UDF that makes predictions.
You might consider the following customizations for your own use case:
- Tune DataLoader's batch_size and num_workers for better performance.
- Use GPU for acceleration.
- Change prediction types.
"""
model = model_fn()
def predict_batch(image_batch):
data_lenght = len(image_batch)
processed_images = np.array([load_and_preprocess_image(img) for img in image_batch])
predictions = model.predict(processed_images, batch_size=BATCH_SIZE)
predicted_labels = [{"class": "Cat", "score": x[0]} if x[0] >= 0.5 else {"class": "Dog", "score": x[0]} for x in predictions]
return pd.DataFrame(predicted_labels)
return_type = "class: string, score:float"
return pandas_udf(return_type, PandasUDFType.SCALAR)(predict_batch)
model_udf = keras_model_udf(cover_model)
sdf.withColumn("prediction", model_udf(col("path"))).show(1)
end_time = time.perf_counter()
return sdf, end_time - start_time
for x in range(len(X1)):
pred, count_time = process_batch_pandas_udf(sdf[x])
X_pudf.append(count_time)
X_pudf_d.append(pred)
print("{} sample: {}s".format(X1[x], count_time))
100 sample: 29.639200400000846s
500 sample: 33.74288650000017s
1000 sample: 38.36985210000057s
1500 sample: 50.09157480000067s
2000 sample: 49.84821749999901s
2500 sample: 46.89816769999743s
3000 sample: 47.90912639999806s
The results are exactly as discussed above when the larger the data, the more pandas udf comes into play.

What next?
Pandas UDFs are a great example of the Spark community’s efforts. Soon I will learn more scala to have a performance comparison of Pandas UDFs with scala. Related article: Instructions for training model Cats vs Dogs Classification using CNN Keras